Workshop
This page gives information on our workshop titled ‘Highly Comparative Analysis of Neural Dynamics’ at CNS2022 on Tuesday 19th July (1:30pm–5pm) at the Melbourne Convention Centre. More information about other workshops is here.

Overview
Quantifying the dynamics of complex, spatially distributed neural systems typically requires researchers to subjectively select from the myriad scientific methods that have been developed to capture properties of univariate dynamics (e.g., properties of the autocorrelation function, multiscale entropy, or thousands of others) and pairwise dependencies (e.g., Pearson correlation, wavelet coherence, or hundreds of others). An emerging alternative to this approach is termed ‘highly comparative time-series analysis’, implemented using associated software tools:
This approach involves simultaneously applying large interdisciplinary libraries of hundreds or thousands of methods to a given dataset, and then extracting informative dynamical patterns from the combined information, or inferring the most informative types of methods through comparison.
This workshop aims to solidify a community around the use of this novel approach and the computational tools for achieving it, and showcase some of highlights of its applications to date. It accompanies an associated CNS2022 tutorial: ‘Characterizing neural dynamics using highly comparative time-series analysis’, which introduces each of these software tools and demonstrates their use on problems involving neural dynamics.
Schedule
- 13:30 Introduction. (Ben Fulcher)
- 13:40 Golia Shafiei: Topographic gradients of intrinsic dynamics across neocortex.
- 14:10 Nao Tsuchiya: Massive feature extraction of EEG data in sleep, anesthesia and perceptual tasks to find the neural correlates of level and contents of consciousness.
- 14:40 Marija Markicevic: Causal effects of microcircuit neuromodulation on resting-state brain dynamics.
- 15:10 Break
- 15:30 Annie Bryant: Characterizing schizophrenia neural dynamics using univariate time-series feature analysis.
- 16:00 Brendan Harris: Inferring parametric variation across non-stationary neural time series.
- 16:30 Angus Leung: Towards blinded classification of levels of consciousness: distinguishing wakefulness from general anesthesia and sleep in flies using a massive library of univariate time-series analyses.
- 17:00 Beers
Speakers and talk information
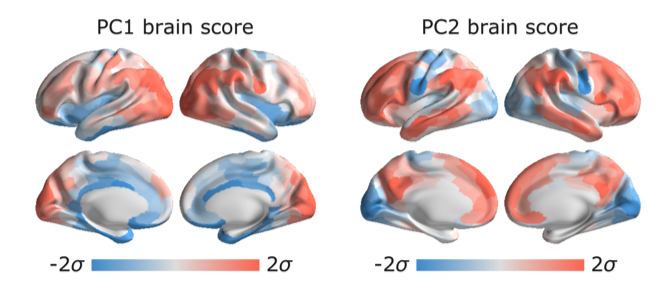
Golia Shafiei (McGill University): Topographic gradients of intrinsic dynamics across neocortex
Neural activity and functional interactions between brain areas are naturally variable from moment to moment, resulting in dynamic configurations of brain activity. Connectome architecture and its spatial embedding shape local and global brain dynamics.
Notably, the functional organization of the human cortex – as well as anatomical markers such as intracortical myelin, cortical thickness, and gene expression – display a hierarchical organization, principally along an axis that spans unimodal sensory/motor and higher order transmodal regions. However, less is known about how regional spontaneous neuronal activity is associated with large-scale network organization.
In this study, we aimed to characterize the link between the dynamic profile of cortical areas and their network embedding using time-series properties of localized brain activity.
We obtained structural and functional magnetic resonance imaging (MRI) data of healthy young adults from the Human Connectome Project.
The highly comparative time series analysis toolbox (hctsa) was used to extract over 7000 temporal features of the resting-state functional MRI time-series. Principal component analysis (PCA) was used to identify linear combinations of time-series features that vary maximally across the cortex. PCA results were compared with gene expression, morphological properties, and functional connectivity of the human cortex. The results show that the dynamic fingerprint of regional brain activity is related to its network embedding.
Local time-series properties are highly similar for pairs of regions that are structurally connected and belong to the same functional networks. These local dynamics recapitulate the unimodal–transmodal hierarchy frequently reported in the human neuroimaging literature.
Altogether, using an unbiased, data-driven approach, the findings demonstrate a dominant hierarchical variation in the properties of spontaneous regional neural activity, providing insight into the dynamical signature of hierarchical functional specialization in the human cortex.
Nao Tsuchiya (Monash University): Massive feature extraction of EEG data in sleep, anesthesia and perceptual tasks to find the neural correlates of level and contents of consciousness
The widely used techniques for EEG data to find the neural correlates of consciousness, be it for the determination of level (e.g., coma, sleep, anesthesia) or contents (e.g., seeing A or B) of consciousness, were traditionally developed based on the visual inspection of electrophysiological recordings by the human eye. Recent computational approaches have complemented these intuitive analyses methods but the choice of analysis tends to be arbitrary. To be agnostic about these prior choice of methods, here we apply hctsa and pyspi methods to EEG data. We will mainly present our completed work on sleep categorization using hctsa but if time permits, we also present our preliminary results on anesthesia with a massive pairwise analysis (pyspi) and event-related ERP data (hctsa) on the distinctions among neural dynamics related to conscious and reported processes, conscious processes in the no-report condition, and non-conscious processes. Our framework lays the groundwork for a data-driven exploration of the neural correlates of consciousness, against which theoretically-derived measures can be compared.
Marija Markicevic (ETH Zurich): Causal effects of microcircuit neuromodulation on resting-state brain dynamics
There is an increasing motivation to understand how the brain’s macroscale dynamics are shaped by underlying microscale mechanisms. In animal models, we can now investigate this relationship in unprecedented detail by directly manipulating cellular-level properties while measuring the whole-brain response using resting-state functional magnetic resonance imaging (rsfMRI). Here, we focused on investigating how cell-specific chemogenetic modulation in either cortical or subcortical regions shapes BOLD fluctuations at the network level. Using ‘highly comparative time-series analysis’ (hctsa), we show the effects of neuromodulation on BOLD time-series dynamics of targeted regions as well as remote but anatomically connected regions. We uncover specific structural and functional constraints that shape changes in BOLD dynamics and illustrate what time-series properties are affected by neuromodulation. Finally, we show that hctsa classifier trained on causal microcircuit alterations can detect similar alterations from an independent rsfMRI dataset of a well-defined disease model. Our results provide deeper insight into how district microcircuit dynamics relate to large-scale noninvasive imaging measurement.
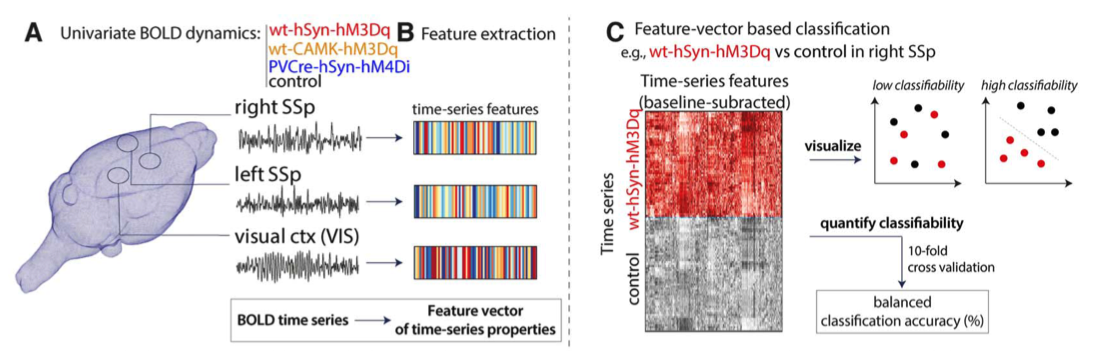
Annie Bryant (Sydney University): Characterizing schizophrenia neural dynamics using univariate time-series feature analysis
Functional neuroimaging offers a window into region-specific neural dysfunction in disorders like schizophrenia. By analysing time-series properties from blood oxygen level-dependent (BOLD) functional magnetic resonance imaging (fMRI), we can infer key regional dynamics that distinguish patients from cognitively healthy controls. Indeed, recent studies comparing BOLD signals in individuals with schizophrenia versus control participants report high classification accuracies, largely using pairwise correlation-based functional connectivity and/or deep learning methods. Despite strong classification metrics, such approaches generally miss region-specific local dynamics and lack biologically interpretable insights due to their intrinsic black-box nature.
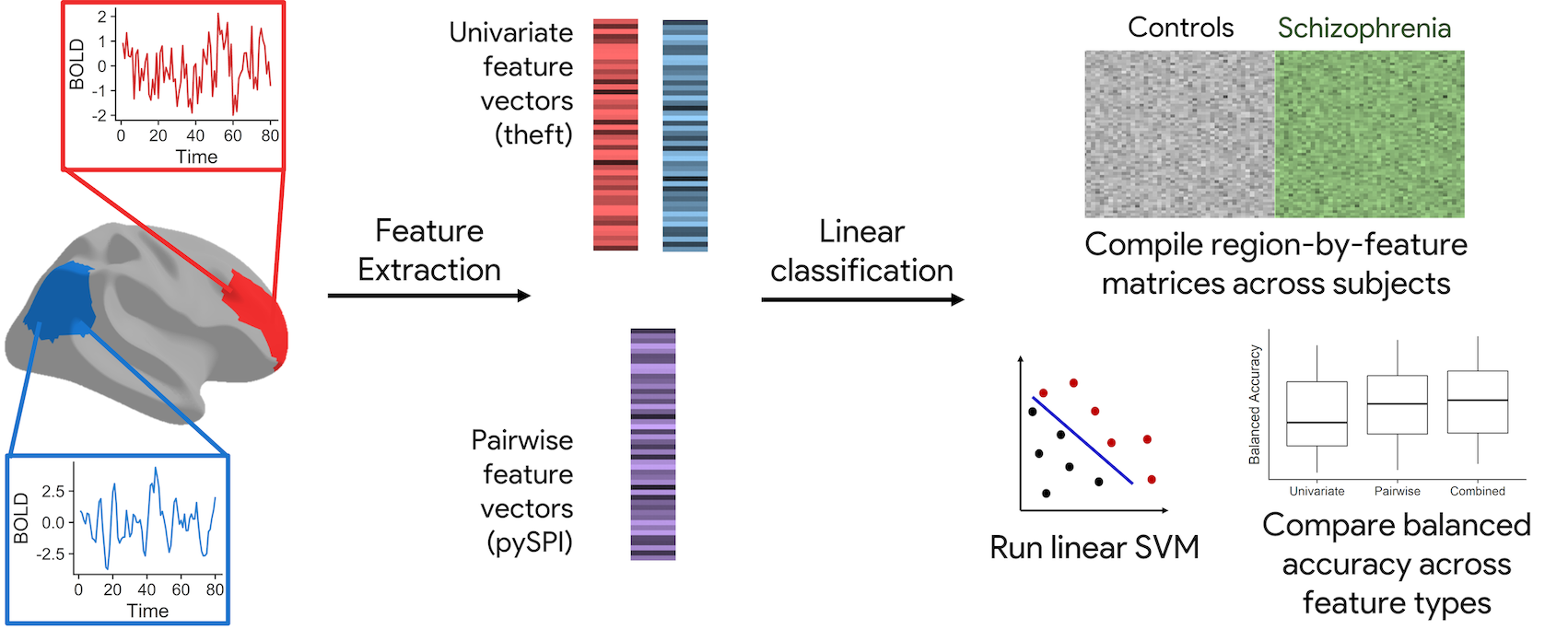
In this talk, I will present a novel approach that integrates local and pairwise neural activity dynamics to classify schizophrenia versus control participants. We will walk through a pipeline that extracts univariate and pairwise features from BOLD fMRI time-series data, using theft (R) and pySPI (python), respectively. We implement a simple linear support vector machine classifier to comprehensively analyze how the time-series features delineate schizophrenia from control participants, with a focus on biological interpretability. We also compare different techniques for dimensionality reduction that can improve out-of-sample classifier robustness. This approach is generalizable across disease spaces and opens the door for deeper insights into the underpinnings of dysregulated neural activity.
Brendan Harris (Sydney University): Inferring parametric variation across non-stationary neural time series
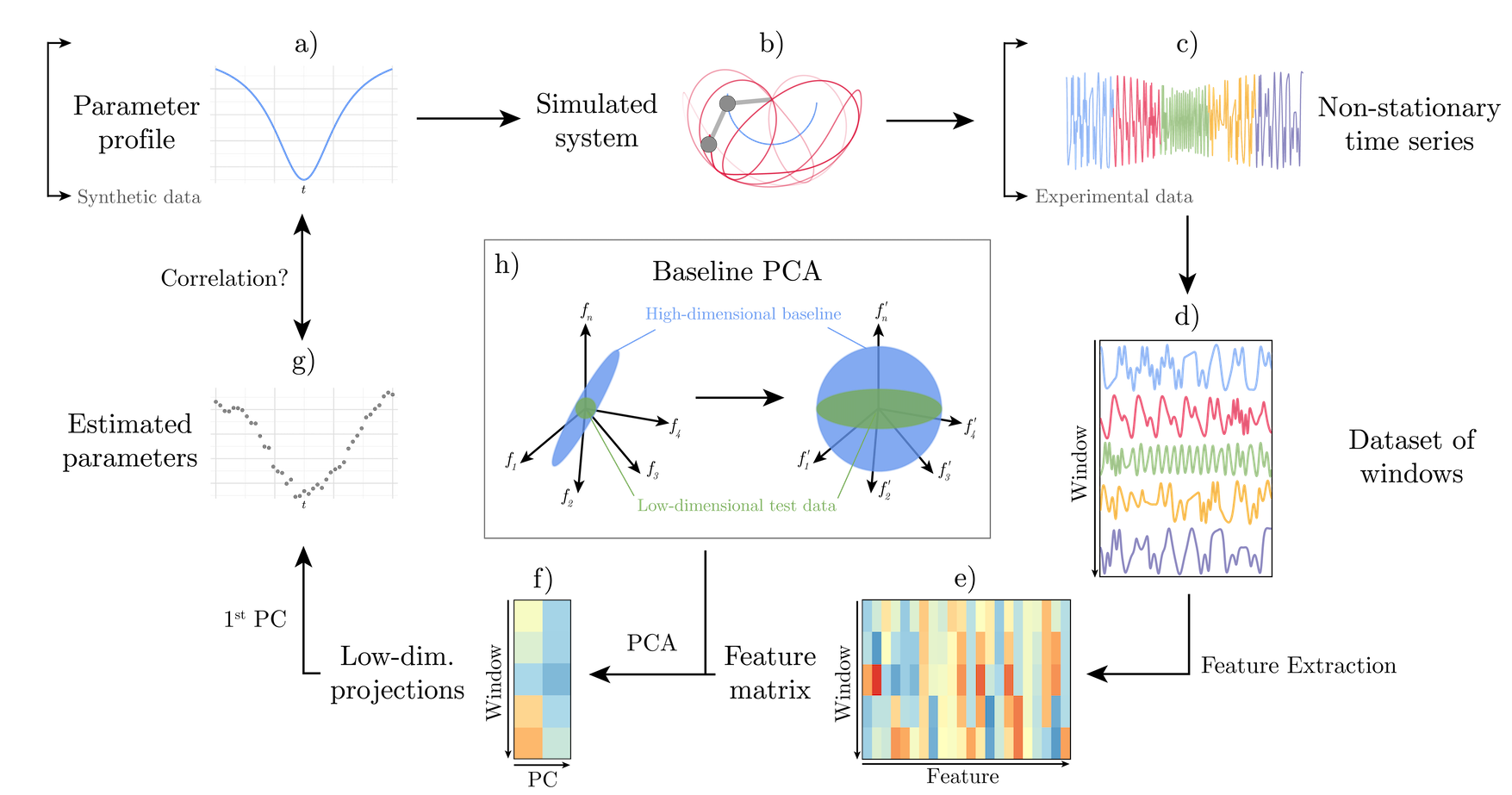
From single-cell neural recordings to waveforms of magnetic fields in the solar wind, non-stationarity in high-dimensional time series often signifies change in the physical parameters of a system. How can we construct intuitive descriptions of arbitrary non-stationarity in time-series data, then use this summary to infer changes in a system’s underlying parameters? In this talk I will present our new method for quantifying non-stationary variation in terms of low-dimensional time-series features, which allows us to understand how the dynamics of a complex system change over time and space. Our method accurately tracks a broad variety of non-stationarities in a range of simulated test systems, where non-stationarity was controlled by ground-truth parameter values. I will also show how we analyse high-dimensional, spatio-temporal Neuropixels recordings to automatically uncover regions of the brain that transition between distinct dynamical states as environmental parameters change over time. Our feature-based approach to dimensionality reduction provides a comprehensive but interpretable summary of non-stationarity that captures meaningful changes in the dynamical properties as well as the underlying parameters of time-varying systems.
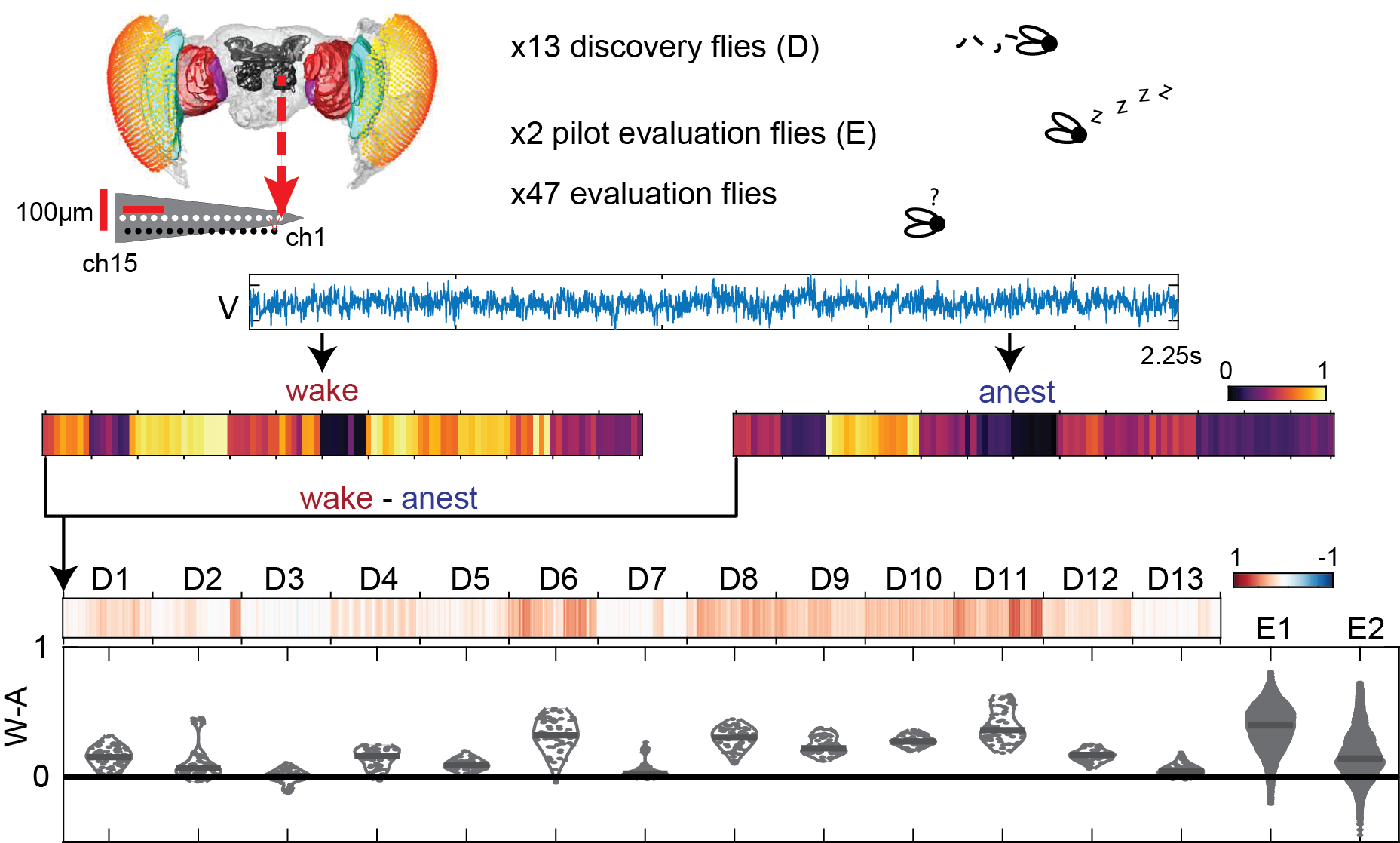
Angus Leung (Monash University): Towards blinded classification of levels of consciousness: distinguishing wakefulness from general anesthesia and sleep in flies using a massive library of univariate time series analyses
The neural mechanisms of consciousness remain elusive. Previous studies on both human and non-human animals, through manipulation of level of conscious arousal, have reported that specific time-series features correlate with level of consciousness, such as spectral power in certain frequency bands. However, such features often lack principled, theoretical justifications as to why they should be related with level of consciousness. This raises two significant issues: firstly, many other types of timesseries features which could also reflect conscious level have been ignored due to researcher biases towards specific analyses; and secondly, it is unclear how to interpret identified features to understand the neural activity underlying consciousness, especially when they are identified from recordings which summate activity across large areas such as electroencephalographic recordings. To address the first concern, here we propose a new approach: in the absence of any theoretical priors, we should be maximally agnostic and treat as many known features as feasible as equally promising candidates. To apply this approach we use highly comparative time-series analysis (hctsa), a toolbox which provides over 7700 different univariate time-series features originating from different research fields. To address the second issue, we employ hctsa to high-quality neural recordings from a relatively simple brain, the fly brain (Drosophila melanogaster), extracting features from local field potentials during wakefulness, general anesthesia and sleep. For each feature, we constructed a classifier for discriminating wakefulness and anesthesia in a discovery group of flies (N = 13).
In this talk, we will assess their performance on a blinded evaluation group of flies (N = 12 for graded levels of anesthesia, N = 18 for single dose anesthesia, and N = 19 for sleep). Pilot results indicate that the performance of only a small subset of features (up to 561, depending on recording location) successfully generalizes to an independent dataset (N = 2). Features which successfully generalize can be fruitful avenues to explore towards robust discoveries of the neural correlates of consciousness.